It is crucial to understand the mechanisms that control the reproductive structures in crops such as maize, as this understanding can greatly improve agricultural productivity. Reproductive structures in plants have a significant impact on crop yield, as they directly influence seed formation and the propagation of crops. Having a comprehensive understanding of the genetic and regulatory factors that contribute to the complexity of reproductive structure evolution can open up opportunities for targeted interventions, optimizing crop yields and overall productivity, especially in the face of a changing climate. My previous work in the Niederhuth lab at Michigan State University, in collaboration with the Monroe lab at the University of California, Davis, has revealed the role of DNA methylation in the evolution of duplicate genes. This research has shown how epigenetically silenced paralogs may accumulate more mutations, potentially leading to pseudogenization or, in some cases, neofunctionalization (Kenchanmane Raju et al. 2023a, 2023b). Kenchanmane-Raju lab is interested in exploring how cell-type expression profiling can help understand changes in paralog expression domains and their role in developmental complexities.
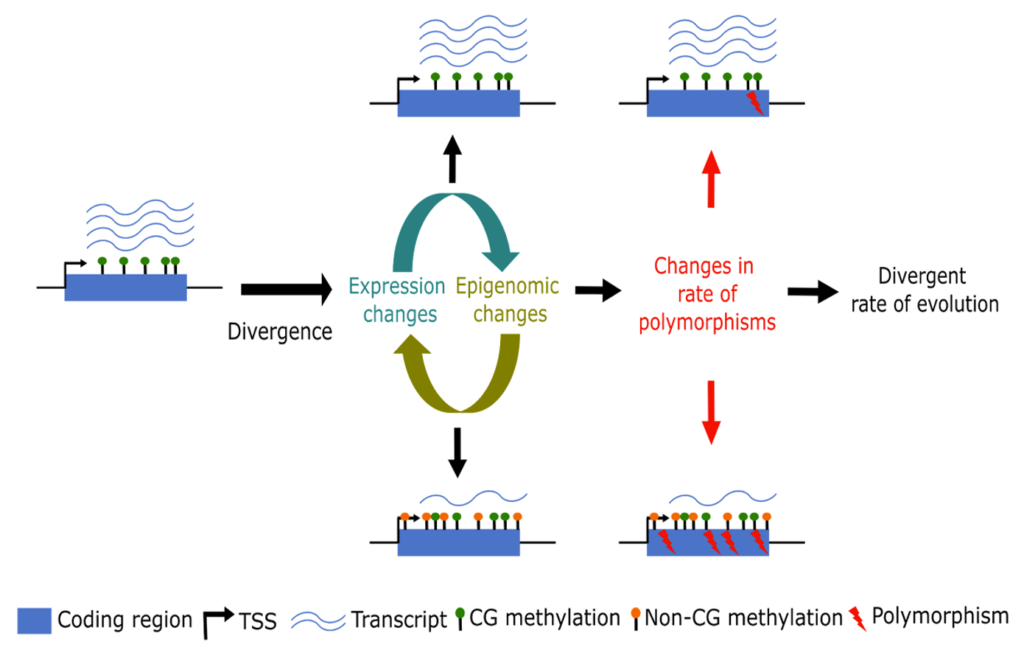
Fig 1: Working model showing differential evolution of duplicate paralogs.