The challenge of climate change is urgent for global agriculture, requiring innovative strategies to enhance crop resilience and ensure food security worldwide. Our previous research, conducted in collaboration with the Schnable lab and Roston lab at the University of Nebraska, has demonstrated the effectiveness of comparative studies in understanding the evolution of chilling stress tolerance in panicoid grasses. These grasses, including maize, sorghum, sugarcane, and millets, are crucial for global food security (Yan et al. 2019; Kenchanmane Raju et al. 2024).

Fig 1. Syntenic orthologs in sorghum and foxtail millet show differential regulation during chilling stress (Kenchanmane Raju et al. 2024)
Developing stress-tolerance traits necessitates a precise understanding of the expression patterns of candidate genes to avoid unintended pleiotropic effects and energy costs. With advancements in single-cell sequencing, researchers can now comprehend gene expression and regulation at the cellular level, which expands our knowledge of plant stress responses. Our previous work in the Birnbaum lab at New York University has illustrated how species-specific traits evolve by utilizing modules from ancestral modules in different cell types (Guillotin et al. 2024).
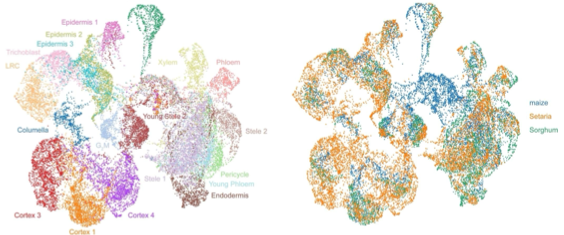
Fig 2. UMAPs generated from integrating single-cell transcriptomes for root tips in maize, sorghum, and setaria (Guillotin et al. 2023)
In summary, comparative single-cell omics can reveal cell-type specific genes that are essential for climate resilience in major crops and offer insights into the evolution of complex traits.